Anamaria Elek
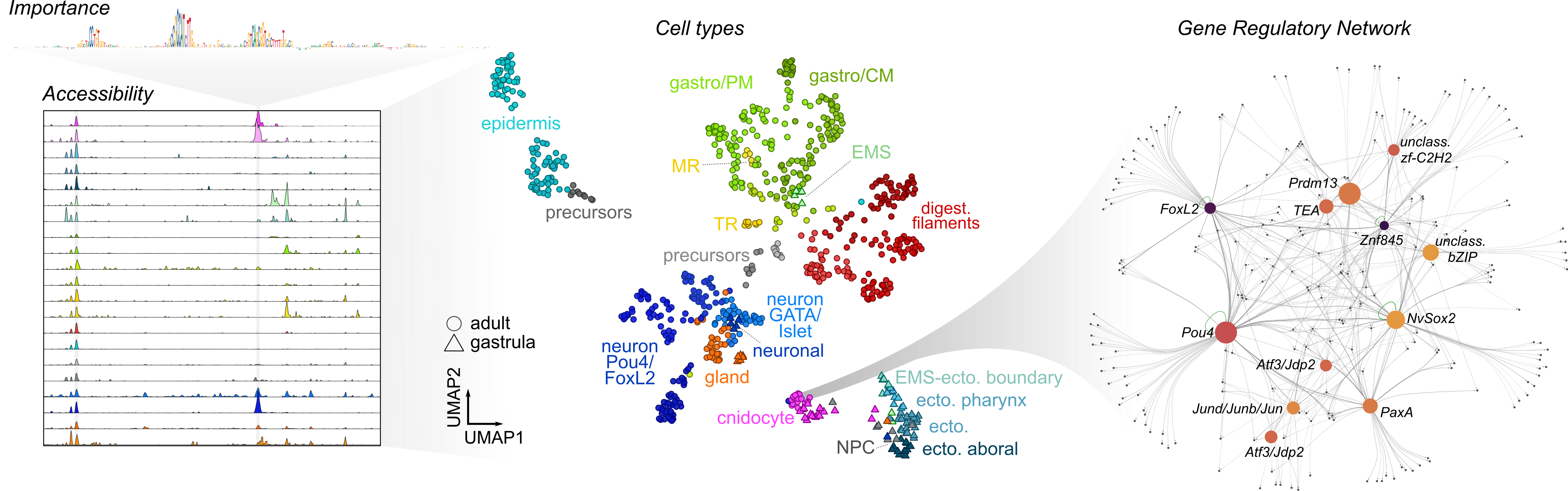
I am a computationl biologist working on understanding gene regulation and expression changes on cell type level, across evolution, development and disease. I have strong foundations in statistics and machine learning, and extensive experience combining data analysis with interactive visualizations that drive meaningful biological discoveries. I obtained my PhD from University Pompeu Fabra, while working at Centre for Genomic Regulation (CRG) in Barcelona, Spain, in the group of Arnau Sebé Pedrós. I was using single cell transcriptomics and chromatin profiling data to dechiper gene regulatory logic behind cell type identities in cnidarians, an early branching group of non-bilaterian animals. During my PhD I also spent several months in the group of Stein Aerts at Center for AI & Computational Biology of VIB-KU Leuven in Leuven, Belgium, where I was training convolutional neural networks for regulatory sequence analysis and cell type comparisons.
Previously, I obtained my Master of Science degree from the University of Zagreb, Croatia, where I worked in the Bioinformatics Group and developed coRdon, a Bioconductor package for the analysis of codon usage. I also worked as a bioinformatician in the Genomics Facility at The Institute of Cancer Research in London, UK.
I am an alumni of NGSchool Society, an NGO that supports bioinformatics community in Central and Eastern Europe. We organized training events for early career stage researchers in computational biology, including a yearly summer school and various conferences, as well as online workshops and lectures. NGSchool is always on the look-out for new people, so if you want to join, don't hesitate to get in touch with them!
I spend my free time running, playing football, hiking, traveling and taking photos.


